Announcing the Genographic Project’s Geno 2.0
In 2005, the National Geographic Society launched the Genographic Project, a first-of-its-kind in-depth look at the deep ancestral genetic patterns of the human race. It was one of the most successful research projects National Geographic has ever undertaken.
Today, National Geographic moves genetic genealogy forward again with the launch of Geno 2.0, a project promising unparalleled and unprecedented breadth and depth of information for human deep ancestry.Dr. Spencer Wells, population geneticist, author of The Journey of Man and other books1 and director of the Genographic Project, announced the launch of the new project and, in a pre-launch presentation to bloggers, provided details on how it came to be and what it will offer.
And for this self-confessed DNA junkie — who never met a DNA test I wouldn’t take — and for anybody with any interest in really understanding human ancestry, this is very exciting stuff.
Wells said planning for Geno 2.0 started nearly two years ago and proceeded from an understanding that technology had advanced far more quickly than the project team could have imagined when it began in 2005. Despite testing more a half-million people between indigenous populations and public participants from more than 130 countries, there was much still unknown about deep human ancestry — and unknowable given the design of the genotyping chips2 then being used for genetic testing.
The first version of the Genographic Project test only looked at a small number of markers of Y-DNA — the DNA passed down a direct paternal line from father to son3 — and of mitochondrial DNA (mtDNA) — the DNA passed down the direct maternal line from a mother to all her children but that only her daughters can pass on.4 It didn’t look at DNA in the rest of the chromosomes — called autosomal DNA or atDNA5 — at all. So, Wells said, the results were “nowhere near today’s cutting edge.”
Getting to that cutting edge wouldn’t be easy. The chips already available weren’t ideal for ancestry. They weren’t fine-tuned enough to get the most information from Y-DNA and mtDNA, but recorded enough medically-relevant information from atDNA to raise the specter of FDA regulation. And the places in the DNA — the markers — sampled by the off-the-shelf chips weren’t chosen for ancestry. For ancestry, you want to look at markers in the DNA that are as distinct as possible among populations, and the existing chips just weren’t designed that way.
So Wells and a team of bioinformatics specialists at Johns Hopkins and other universities designed an all-new chip from the ground up. It’s manufactured by Illumina, and results will be processed at the Family Tree DNA lab in Houston, Texas. This new chip will test all three kinds of DNA — Y-DNA, mtDNA and atDNA — and in ways nobody has ever done before.First and foremost, the chip is specifically designed to avoid sampling any known medically relevant markers. “A major purpose in designing this genotyping platform was to ensure that we did not bring Big Brother out of the woodwork. Government regulation of ancestral testing would be a catastrophe,” Wells said. If there was an error in choosing SNPs, it was one was of overexclusion. “We want to be sure that the information we gather can’t be exploited for purposes other than an interest in one’s own ancestry.”
For Y-DNA, the chip looks at more than 12,000 SNPs — pronounced snips and meaning single-nucleotide polymorphisms, a type of DNA sequence variation6 — the majority of which are as yet unpublished. In doing so, the chip “has re-defined the Y-chromosome tree in significant ways.” For mtDNA, the chip looks at roughly 3,200 unique SNPs. As a result, Wells said, the test offers “more resolution that any other currently available test for Y-DNA and mtDNA.”
Functionally it replaces any existing Y-DNA or mtDNA deep clade test, and this is a really big deal in genealogy. Knowing exactly where our DNA falls in terms of haplogroup down to the nth degree is what helps us figure out if we’re from this branch of the family tree or that one, and this test is going to move the ball way down field for both Y-DNA and mtDNA and for both paternal and maternal lines.
Just as exciting is what the test promises to tell us about just what branch of the human family tree we’re from. The SNPs selected from the atDNA and X chromosomal DNA were chosen to emphasize and focus on what are called Ancestry Informative Markers, or AIMs. Those are markers that show more of the differences among populations and so are extremely useful in distinguishing them.7 Wells explained: “Most populations can be distinguished even when closely related, such as Syrian and Druze. The more fine-tuned the AIMs are, the more accurate we can be in identifying populations. We won’t be saying European. Eventually we might be saying Finnish or Austrian or German.” And the atDNA segments are also aimed at sampling more of the pre-human genetic components than ever before, including Neanderthal and Denisovan.
Overall, this part of the test is designed to complement, not replace, existing autosomal tests like Family Finder from Family Tree DNA or Relative Finder from 23andMe. It’s aimed not at finding cousins but rather at disclosing our deep ancestral admixture.
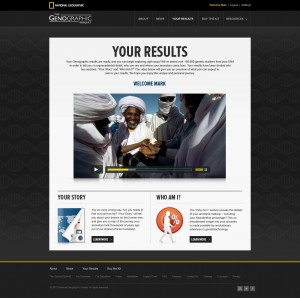
Test results will be displayed in an entirely new way with segments including Your Story (information on haplogroups and the journey ancestors took out of Africa), Who Am I? (details of ancestral makeup including regional genetic affiliations, ancient populations such as Neanderthal percentage and closest population today), and a community feature shown as a unique circular display of matches in the database where you can interact through the Geno 2.0 website to share genealogical information.Only one cheek-swab kit will be needed for males to get all their results — Y-DNA, mtDNA, and atDNA. Females will get their mtDNA and atDNA results, and they’ll be able to link to a male relative to display their paternal line Y-DNA in their results. Data from Geno 2.0 can be integrated with the Family Tree DNA database at the option of the person tested at no cost. The link between Geno 2.0 and Family Tree DNA should open up sometime in the early fall.
People who’ve already tested with Family Tree DNA will have the option in the future to have any existing sample tested with Geno 2.0 at an as-yet-unspecified discount — a link will appear on the person’s FTDNA personal page later this year if the person is eligible to participate this way. (UPDATE (22 Aug 2012): FTDNA has backed off from the possibility of this option. A recent announcement from FTDNA simply referred folks to the Geno 2.0 site.) Integration isn’t possible with other companies’ tests because of the unique collection of markers on the chip, but the data is compatible with existing test data from all companies and raw data can be downloaded for use with third-party utilities.
The cost of testing with Geno 2.0 is $199.95 with free USA shipping, and kits are available now for pre-order at the Geno 2.0 website at www.genographic.com. The first kits are expected to go out in early September — including, truth in blogging here, a free one for yours truly — and the estimated time to results is six to eight weeks after the cheek swabs are returned. No waiting list is expected, Wells said. “We’re prepared for as many people as want to order.”
More information on Geno 2.0 is available at at the Geno 2.0 website, and more will be disclosed in the weeks and months to come. Wells promised that Geno 2.0, like its predecessor, will have a broad project reach. And for other viewpoints and discussions about this announcement, here are other blogs where this is sure to be a hot topic:
• Blaine Bettinger, The Genetic Genealogist
• CeCe Moore, Your Genetic Genealogist
Sigh. More DNA goodies. I can’t wait…
SOURCES
- See Spencer Wells, The Journey of Man: A Genetic Odyssey (Princeton, N.J. : Princeton University Press, 2002); Spencer Wells, Deep Ancestry: Inside The Genographic Project (Washington, D.C. : National Geographic, 2006); Spencer Wells, Pandora’s Seed: The Unforeseen Cost of Civilization (New York : Random House, 2010). ↩
- These are small biotechnological chips that can “read” a DNA sample. For an overview of genotyping chips, see Applications / Genotyping, Illumina (http://www.illumina.com : accessed 25 Jul 2012). ↩
- ISOGG Wiki (http://www.isogg.org/wiki), “Y chromosome DNA test,” rev. 26 Jun 2012. ↩
- ISOGG Wiki (http://www.isogg.org/wiki), “Mitochondrial DNA,” rev. 30 Jul 2010. ↩
- ISOGG Wiki (http://www.isogg.org/wiki), “Autosomal DNA,” rev. 8 Feb 2012. ↩
- ISOGG Wiki (http://www.isogg.org/wiki), “Single-nucleotide polymorphism,” rev. 1 Feb 2012. ↩
- ISOGG Wiki (http://www.isogg.org/wiki), “Ancestry-informative markers,” rev. 14 Jul 2010. ↩





Nice job, as always, Judy!
Thanks, CeCe! Love the detail in your report as well — so MUCH information here!
From one DNA junkie to another [do you suppose that’s our common DNA?]
This is exciting. I can’t wait to see the link between FT DNA and Geno 2.0!
Definitely exciting, Kay. It’s going to tell us more about deep human ancestry than we’ve ever known before. Cool stuff!